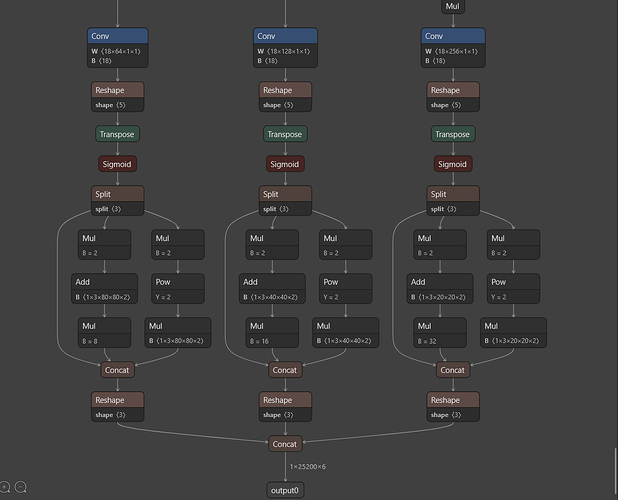
I was running into a pretty much identical error, and suspect others may too if they’re using yolo5 with DFC
My parsing step is as follows, with the start/end nodes based on what I found when opening in netron.app - you’ll find these final conv nodes right at the end of the model:
runner = ClientRunner(hw_arch=chosen_hw_arch)
hn, npz = runner.translate_onnx_model(
onnx_path,
onnx_model_name,
start_node_names=["/model.0/conv/Conv"],
end_node_names=["/model.24/m.2/Conv",
"/model.24/m.1/Conv",
"/model.24/m.0/Conv"],
net_input_shapes={"/model.0/conv/Conv": [1, 3, 640, 640]},
)
my alls script looks like:
alls = """
normalization1 = normalization([0.0, 0.0, 0.0], [255.0, 255.0, 255.0])
change_output_activation(sigmoid)
nms_postprocess("/local/shared_with_docker/visdrone/postprocess_config/yolov5s_nms_config.json", yolov5, engine=cpu)
model_optimization_config(calibration, batch_size=1)
"""
I’ve pull this from the hailomz repo and edited only the nms path. The nms config I’ve copied from the repo and left unchanged
The original model was obtained by running the docker image, then running a simple:
python train.py --img 640 --batch 16 --epochs 300 --data coco128.yaml --weights yolov5s.pt --cfg models/yolov5s.yaml
python export.py --data=coco128.yaml --weights=yolov5s.pt --opset=14 --include onnx
However I wa getting ths ‘layer not found’ error when I ran:
runner.load_model_script(alls)
runner.optimize_full_precision()
Layer 63 is clearly used for box decoding in the nms file I linked above. I did’t understand why this isn’t found in my model
Though I recalled a guide for yolo11 and dfc here, they helpfully recommended using this to inspect your parsed model, because the nodes will get renamed. And indeed, when I checked for the original names of my end nodes, they’d been renamed. I needed to amend the nms_config file to match these layers:
from hailo_sdk_client import ClientRunner
# Load the HAR file
har_path = hailo_model_har_name
runner = ClientRunner(har=har_path)
from pprint import pprint
try:
# Access the HailoNet as an OrderedDict
hn_dict = runner.get_hn() # Or use runner._hn if get_hn() is unavailable
print("Inspecting layers from HailoNet (OrderedDict):")
# Pretty-print each layer
for key, value in hn_dict.items():
print(f"Key: {key}")
pprint(value)
print("\n" + "="*80 + "\n") # Add a separator between layers for clarity
except Exception as e:
print(f"Error while inspecting hn_dict: {e}")
This yields the entire model dict, an example of which here:
('yolo5s/conv60',
OrderedDict([('type', 'conv'),
('input', ['yolo5s/conv59']),
('output', ['yolo5s/output_layer3']),
('input_shapes', [[-1, 20, 20, 512]]),
('output_shapes', [[-1, 20, 20, 255]]),
('original_names', ['/model.24/m.2/Conv']),
('compilation_params', {}),
('quantization_params', {}),
('params',
OrderedDict([('kernel_shape', [1, 1, 512, 255]),
('strides', [1, 1, 1, 1]),
('dilations', [1, 1, 1, 1]),
('padding', 'VALID'),
('groups', 1),
('layer_disparity', 1),
('input_disparity', 1),
('batch_norm', False),
('elementwise_add', False),
('activation', 'linear')]))])),
here you’ll see that one of my final output layers is actually now called yolo5s/conv60 - yet this is absent in the nms_config file (which instead had conv63 listed, a layer I couldn’t see in the dict)
The fix was simple in the end, just map the output conv nodes to the ‘new name’, then ensure the nms file matches these. For example, in the case of this conv60 layer, I updated my nms as such:
{
"name": "bbox_decoder60",
"w": [
116,
156,
373
],
"h": [
90,
198,
326
],
"stride": 32,
"encoded_layer": "conv60"
}
This seems to work as expected, I optimized the model and it has valid predictions. My question for @omria is why the yolo5s nms config file in the repo has different box decoder layer names to this yolo5s model I’ve used (since it wasn’t customised on any new data or amended otherwise)?
full error message which might help direct others here, since it doesn’t seem like a terribly common problem
---------------------------------------------------------------------------
HailoNNException Traceback (most recent call last)
Cell In [36], line 8
1 alls = """
2 normalization1 = normalization([0.0, 0.0, 0.0], [255.0, 255.0, 255.0])
3 change_output_activation(sigmoid)
4 nms_postprocess("/local/shared_with_docker/visdrone/postprocess_config/yolov5s_nms_config.json", yolov5, engine=cpu)
5 model_optimization_config(calibration, batch_size=1)
6 """
7 runner.load_model_script(alls)
----> 8 runner.optimize_full_precision()
10 # runner.optimize(calib_dataset)
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_common/states/states.py:16, in allowed_states.<locals>.wrap.<locals>.wrapped_func(self, *args, **kwargs)
12 if self._state not in states:
13 raise InvalidStateException(
14 f"The execution of {func.__name__} is not available under the state: {self._state.value}",
15 )
---> 16 return func(self, *args, **kwargs)
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_client/runner/client_runner.py:2031, in ClientRunner.optimize_full_precision(self, calib_data, data_type)
2022 if calib_data is None and not (
2023 self._sdk_backend.calibration_data or self._sdk_backend.calibration_data_random_max
2024 ):
2025 self._logger.deprecation_warning(
2026 "Optimizing in full precision will require calibration data "
2027 "in the near future, to allow more accurate optimization "
2028 "algorithms which require inference on actual data.",
2029 DeprecationVersion.JUL2024,
2030 )
-> 2031 self._optimize_full_precision(calib_data=calib_data, data_type=data_type)
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_client/runner/client_runner.py:2034, in ClientRunner._optimize_full_precision(self, calib_data, data_type)
2033 def _optimize_full_precision(self, calib_data=None, data_type=None):
-> 2034 self._sdk_backend.optimize_full_precision(calib_data=calib_data, data_type=data_type)
2035 self._state = States.FP_OPTIMIZED_MODEL
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_client/sdk_backend/sdk_backend.py:1591, in SDKBackendQuantization.optimize_full_precision(self, update_model_and_params, calib_data, data_type)
1588 params = ModelParams({})
1589 params.set_params_kind(ParamsKinds.FP_OPTIMIZED)
-> 1591 model, params = self._apply_model_modification_commands(model, params, update_model_and_params)
1592 if calib_data is None:
1593 self._logger.debug("Use random calibration data for full precision optimization")
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_client/sdk_backend/sdk_backend.py:1482, in SDKBackendQuantization._apply_model_modification_commands(self, model, params, update_model_and_params)
1474 self._logger.warning(
1475 "Be advised the Dataflow Compiler default YUV to RGB/BGR conversion uses YUV601 standard, which is different than ISP and may cause accuracy degradation."
1476 "For better results consider using yuv_full_range_to_rgb/bgr instead."
1477 )
1478 self._logger.warning(
1479 "The Dataflow Compiler default YUV to RGB/BGR will be changed to yuv_full_range_to_rgb/bgr in the next version.",
1480 )
-> 1482 model, params = command.apply(model, params, hw_consts=self.hw_arch.consts)
1483 if update_model_and_params:
1484 for layer_name in command.meta_data:
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_client/sdk_backend/script_parser/nms_postprocess_command.py:397, in NMSPostprocessCommand.apply(self, hailo_nn, params, hw_consts)
391 if self.engine == PostprocessTarget.NN_CORE.value:
392 logger.warning(
393 "Currently `nn-core` performs only score threshold. In the near future it will perform score"
394 " + IoU threshold. If you want to apply IoU threshold please use `cpu` or `auto` instead",
395 )
--> 397 self._update_config_file(hailo_nn)
398 self.validate_nms_config_json()
399 pp_creator = create_nms_postprocess(
400 hailo_nn,
401 params,
(...)
408 self.dfl_on_nn_core,
409 )
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_client/sdk_backend/script_parser/nms_postprocess_command.py:557, in NMSPostprocessCommand._update_config_file(self, hailo_nn)
555 self._update_config_file_by_engine()
556 self._update_command_args_in_config_file()
--> 557 self._layers_scope_addition(hailo_nn)
558 self._update_config_layers(hailo_nn)
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_client/sdk_backend/script_parser/nms_postprocess_command.py:600, in NMSPostprocessCommand._layers_scope_addition(self, hailo_nn)
598 scope_addition_fields = [field for field in fields_with_layer_name if bbox_decoder.get(field.value)]
599 for field in scope_addition_fields:
--> 600 bbox_decoder[field.value] = hailo_nn.get_layer_by_name(bbox_decoder[field.value]).name
File /local/workspace/hailo_virtualenv/lib/python3.10/site-packages/hailo_sdk_common/hailo_nn/hailo_nn.py:537, in HailoNN.get_layer_by_name(self, layer_name)
532 if len(potential_matches) > 1:
533 raise HailoNNException(
534 f"The layer named {layer_name} exist under multiple scopes in the HN {[layer.scope for layer in potential_matches]}",
535 )
--> 537 raise HailoNNException(f"The layer named {layer_name} doesn't exist in the HN")
HailoNNException: The layer named conv63 doesn't exist in the HN